TL;DR.
CryoFM is a generative foundation model for cryo-EM density reconstruction. It learns a generalizable structural prior from thousands of high-quality cryo-EM maps in an unsupervised manner and combines it with explicit experimental likelihoods for Bayesian inference. This enables controllable posterior sampling for denoising, restoration, and refinement.
Overview
Single-particle cryo-EM density reconstruction is a severely ill-posed inverse problem, commonly degraded by noise, preferred orientations, and reconstruction artifacts. Existing approaches either rely on strong hand-crafted assumptions or supervised post-processing models, which can be brittle and prone to hallucinating structural features.
CryoFM addresses this challenge by introducing a generative foundation model for cryo-EM densities. The model is trained unsupervised on thousands of high-quality cryo-EM maps using flow matching, learning a reusable prior over macromolecular density distributions that generalizes across molecular systems.
At inference time, this learned prior is combined with explicit likelihood models that describe experimental degradations, forming a principled Bayesian inference framework. Within this framework, cryoFM performs flow-based posterior sampling, an inference-only procedure that simultaneously enables denoising, restoration, and refinement while remaining explicitly constrained by dataset-derived statistics.
CryoFM consistently improves density reconstruction across diverse and challenging experimental settings, including datasets with strong preferred orientations and spatially heterogeneous signal-to-noise ratios, without introducing hallucinated features. In addition, the model can be fine-tuned into conditional generative models for density post-processing, producing maps with improved interpretability and fewer artifacts compared to existing supervised methods.
Overall, cryoFM establishes generative foundation models as a principled, controllable, and physically grounded framework for cryo-EM density reconstruction and modification.
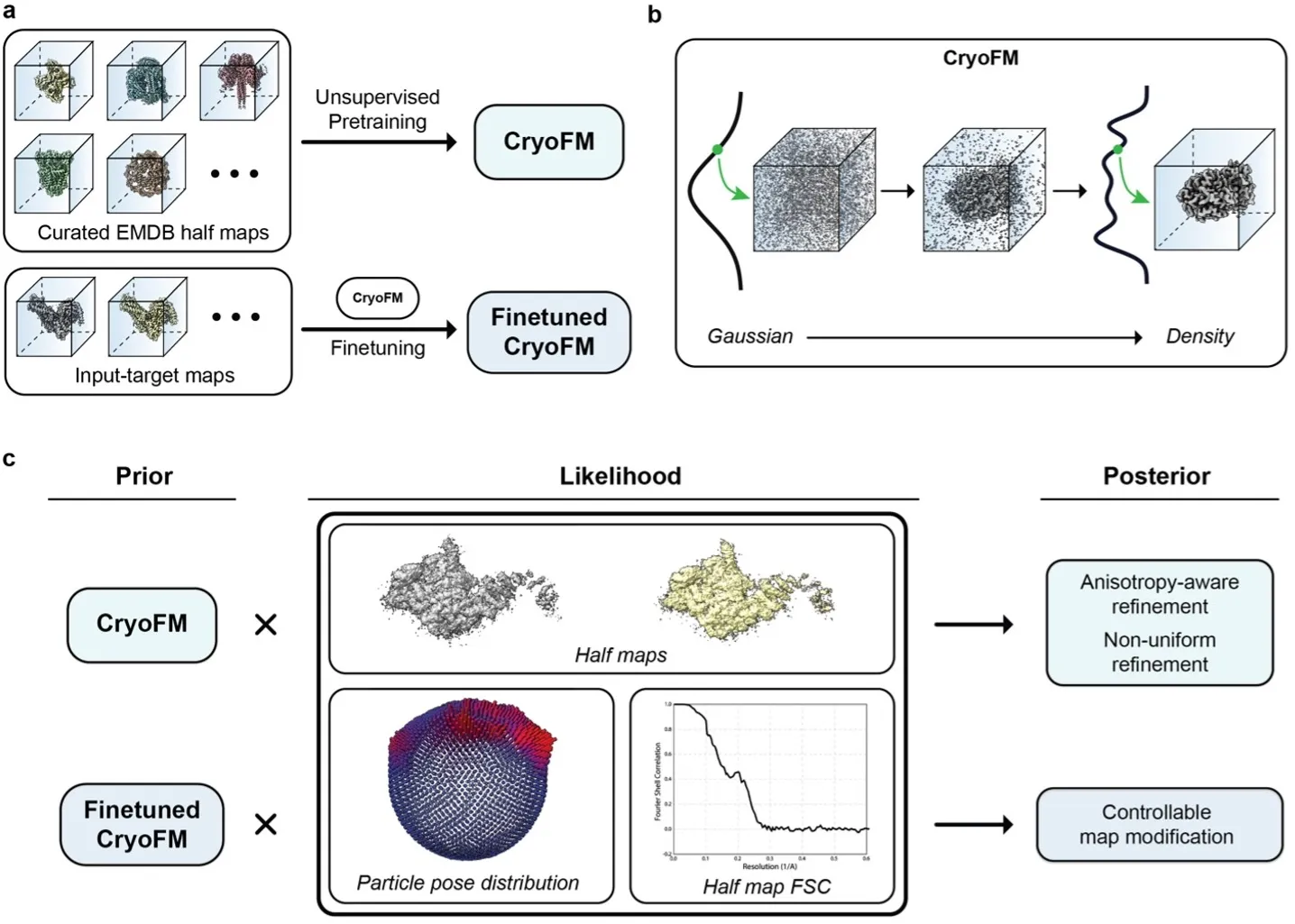
CryoFM is a generative foundation model learns the distribution of cryo-EM densities and enables various downstream tasks.
What cryoFM can do?
Data-constrained generative inference with cryoFM
CryoFM enables data-constrained generative inference for cryo-EM densities by combining a learned generative prior with explicit likelihood models. A flow-matching–trained prior transforms samples from a simple Gaussian distribution into realistic cryo-EM density maps, while dataset-specific likelihood terms guide inference toward solutions consistent with the observed data. During posterior sampling, the learned prior flow captures structural regularities, and likelihood-driven corrections enforce experimental constraints.
We demonstrate cryoFM on both denoising and anisotropy correction tasks using synthetic benchmarks. For denoising, input densities are synthetically corrupted and subsequently restored using cryoFM, with both inputs and outputs post-processed using identical FSC-based filtering and B-factor sharpening for fair comparison. Quantitative evaluation using FSC curves shows consistent improvement of the cryoFM-denoised densities relative to the noisy inputs.
To evaluate anisotropy correction, directional masks are applied in Fourier space to simulate missing information caused by anisotropic sampling. CryoFM successfully restores masked frequency regions, producing reconstructions that closely match the ground-truth densities. Compared with existing restoration methods, cryoFM achieves higher FSC scores within the masked Fourier-space regions, demonstrating its ability to recover missing structural information while remaining constrained by the data.
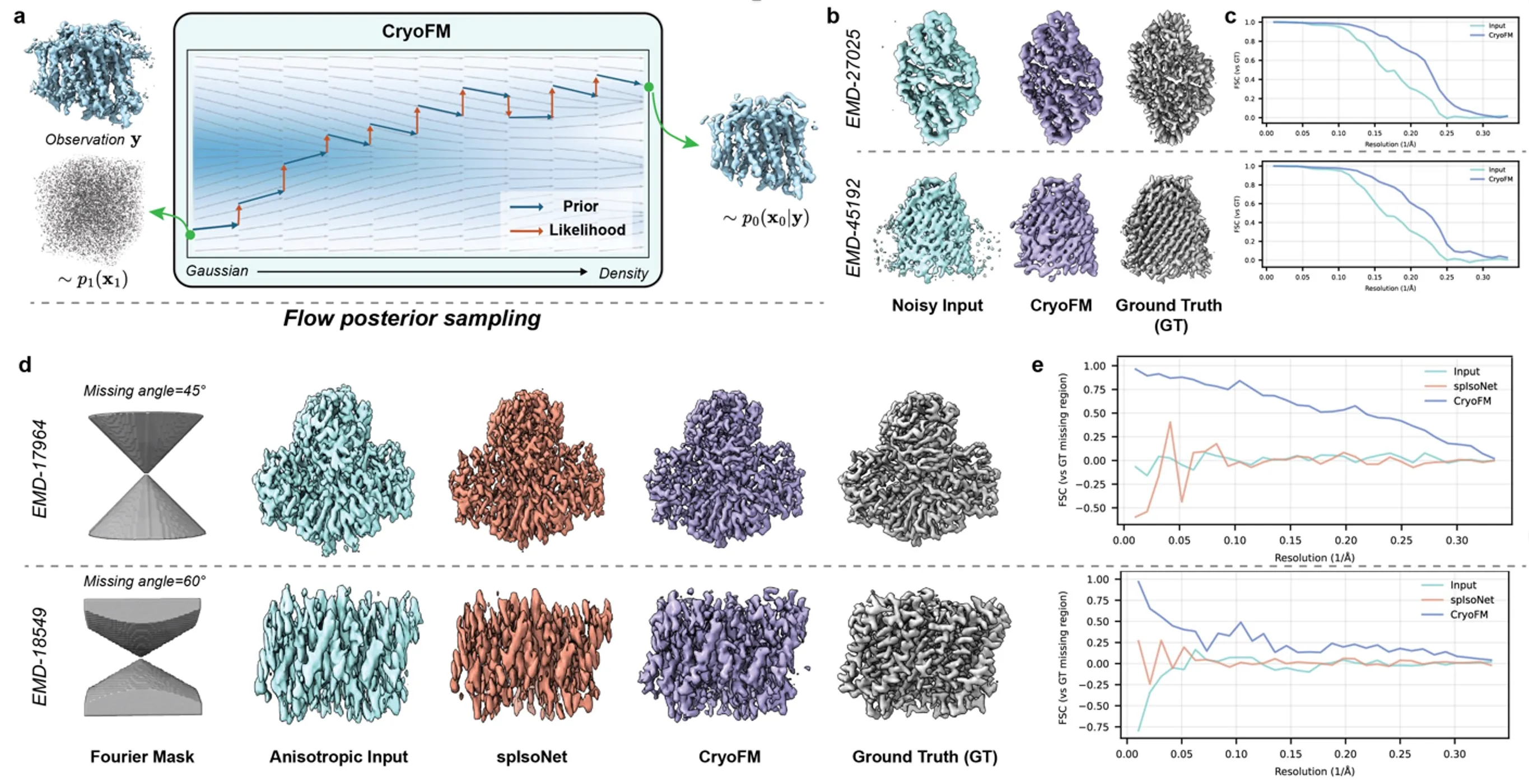
CryoFM enables data constrained inference by posterior sampling.
Anisotropy-aware refinement with cryoFM
cryoFM can be integrated into standard cryo-EM refinement pipelines to perform anisotropy-aware density refinement. When combined with the RELION expectation–maximization (EM) framework, cryoFM performs flow-based posterior sampling guided by likelihoods derived from particle pose distributions, explicitly mitigating reconstruction artifacts caused by anisotropic sampling.
We evaluate this approach on real experimental datasets from EMPIAR-10096 and EMPIAR-10097. Compared to standard RELION reconstructions, cryoFM-based refinement consistently produces density maps with improved local resolution and clearer structural features, as visualized using local resolution coloring.
Quantitative evaluation using model–map FSC further confirms these improvements. Across a broad resolution range, cryoFM exhibits higher FSC values than standard RELION, demonstrating that anisotropy-aware generative refinement leads to both visually and quantitatively superior reconstructions.
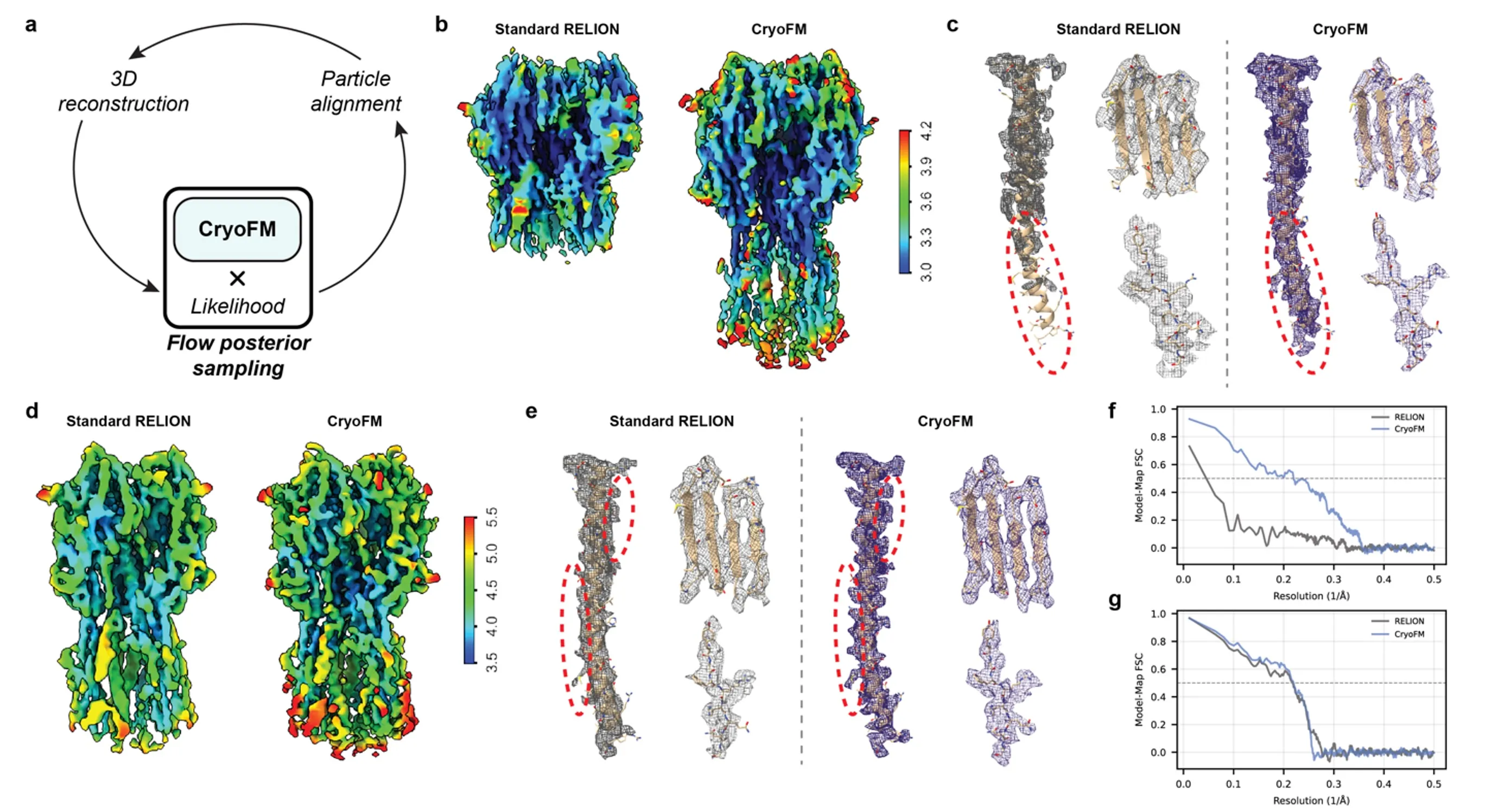
Anisotropy-aware refinement.
Non-uniform refinement with cryoFM across benchmark datasets
CryoFM supports non-uniform density refinement that improves reconstruction quality across a wide range of benchmark cryo-EM datasets. We compare standard RELION reconstructions with cryoFM non-uniform refinement on three representative datasets (EMPIAR-10330, EMPIAR-11762, and EMPIAR-12510).
Across all datasets, cryoFM produces density maps with consistently improved local resolution, as visualized using local-resolution coloring. Zoomed-in views of representative regions show sharper density and improved agreement with docked atomic models, including more clearly resolved side-chain features. These improvements highlight qualitative gains that go beyond global resolution metrics.
Quantitative evaluation using half-map FSC further confirms the advantage of cryoFM. Across a broad resolution range, cryoFM achieves higher FSC values than standard RELION, demonstrating robust and consistent improvements in both global and local reconstruction quality.
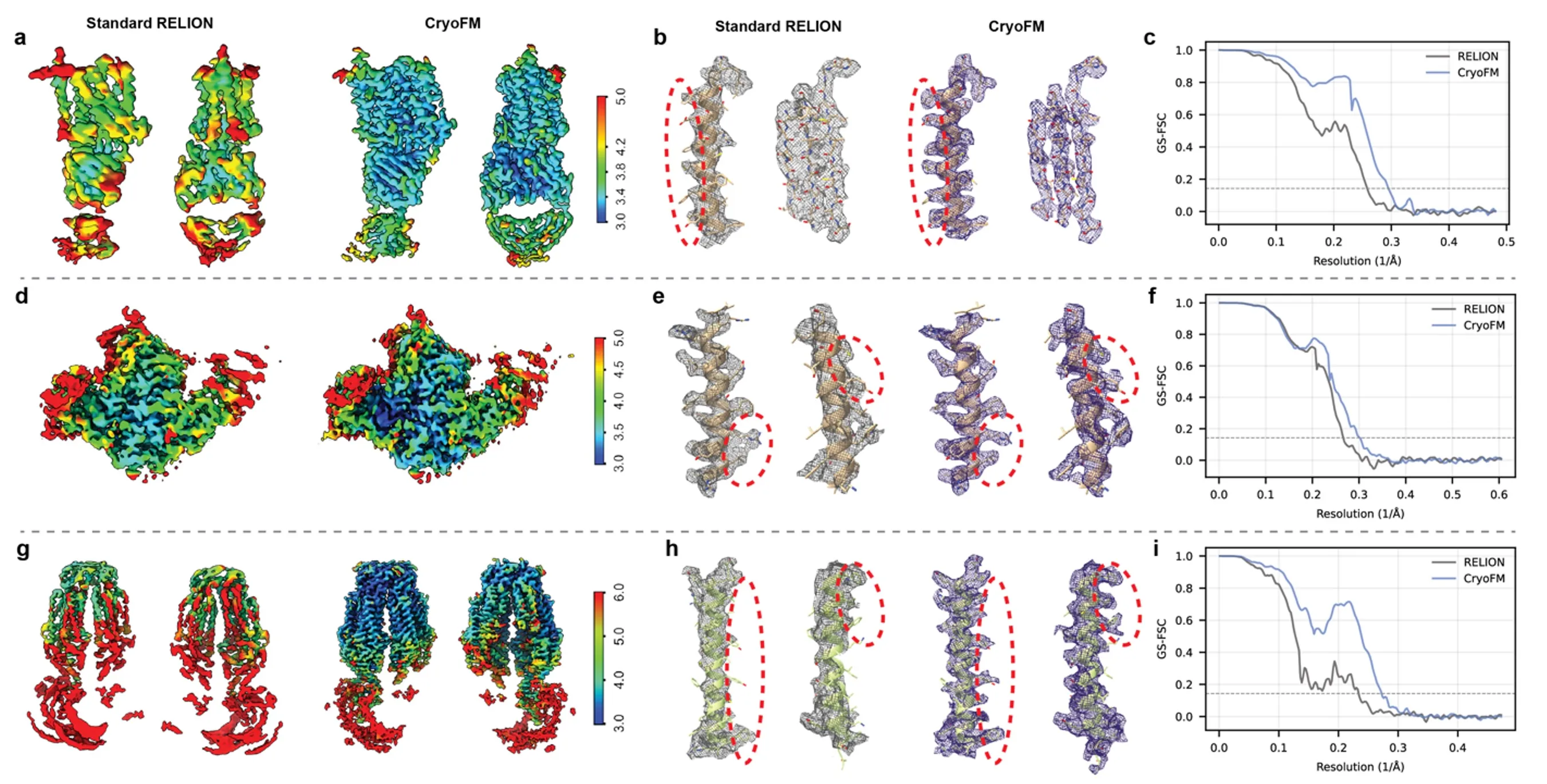
Non-uniform refinement.
Controllable density post-processing with cryoFM
CryoFM enables controllable density post-processing through fine-tuning and likelihood-guided generation. By fine-tuning the pretrained foundation model on paired input–target density maps, cryoFM can adapt to different post-processing styles while retaining the general structural prior learned during pretraining.
We evaluate this capability by comparing cryoFM with existing supervised post-processing methods, including DeepEMhancer1 and EMReady2. Quantitative evaluation using map–model FSC at the 0.5 cutoff shows that fine-tuned cryoFM consistently achieves competitive or improved resolution compared to both the original input maps and existing post-processing approaches.
Qualitative comparisons further highlight the differences in output style. cryoFM produces density maps with improved interpretability and clearer structural features, while avoiding excessive sharpening or artificial texture. Importantly, the generated densities remain consistent with the underlying experimental data.
Beyond standalone post-processing, fine-tuned cryoFM can also be used as a generative prior combined with task-specific likelihoods. This enables likelihood-guided posterior sampling for controllable density modification, such as EMReady-style enhancement applied directly within an anisotropy-aware reconstruction framework.
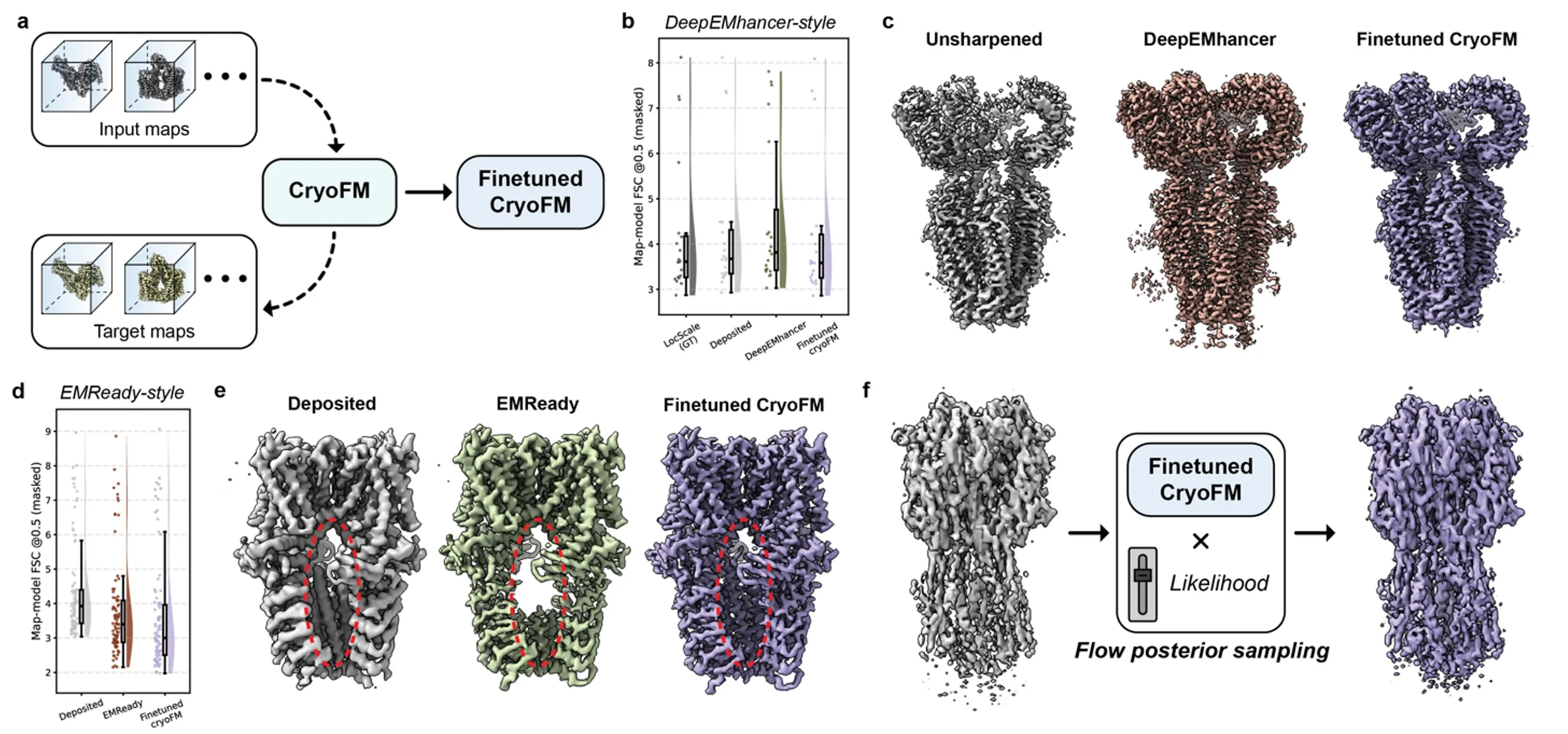
Controllable density post-processing.
Citation
If you find CryoFM2 helpful, please cite:
@article {Li2025.12.29.696802,
author={Li, Yilai and Yuan, Jing and Zhou, Yi and Wang, Zhenghua and Chen, Suyi and Yang, Fengyu and Ling, Haibin and Kovalsky, Shahar Z and Zheng, Xiaoqing and Gu, Quanquan},
title={A Generative Foundation Model for Cryo-EM Densities},
elocation-id={2025.12.29.696802},
year={2025},
doi={10.64898/2025.12.29.696802},
publisher={Cold Spring Harbor Laboratory},
URL={https://www.biorxiv.org/content/early/2025/12/29/2025.12.29.696802},
eprint={https://www.biorxiv.org/content/early/2025/12/29/2025.12.29.696802.full.pdf},
journal={bioRxiv}
}Footnotes
-
DeepEMhancer is introduced in “DeepEMhancer: a deep learning solution for cryo-EM volume post-processing” by Ruben Sanchez-Garcia et al., 2021; see Communications Biology. ↩
-
EMReady is introduced in “Improvement of cryo-EM maps by simultaneous local and non-local deep learning” by Jiahua He et al., 2023; see Nature Communications. ↩