CryoFM2: Overview
A Generative Foundation Model for Cryo-EM Densities
under review, 2025.
What is CryoFM2?
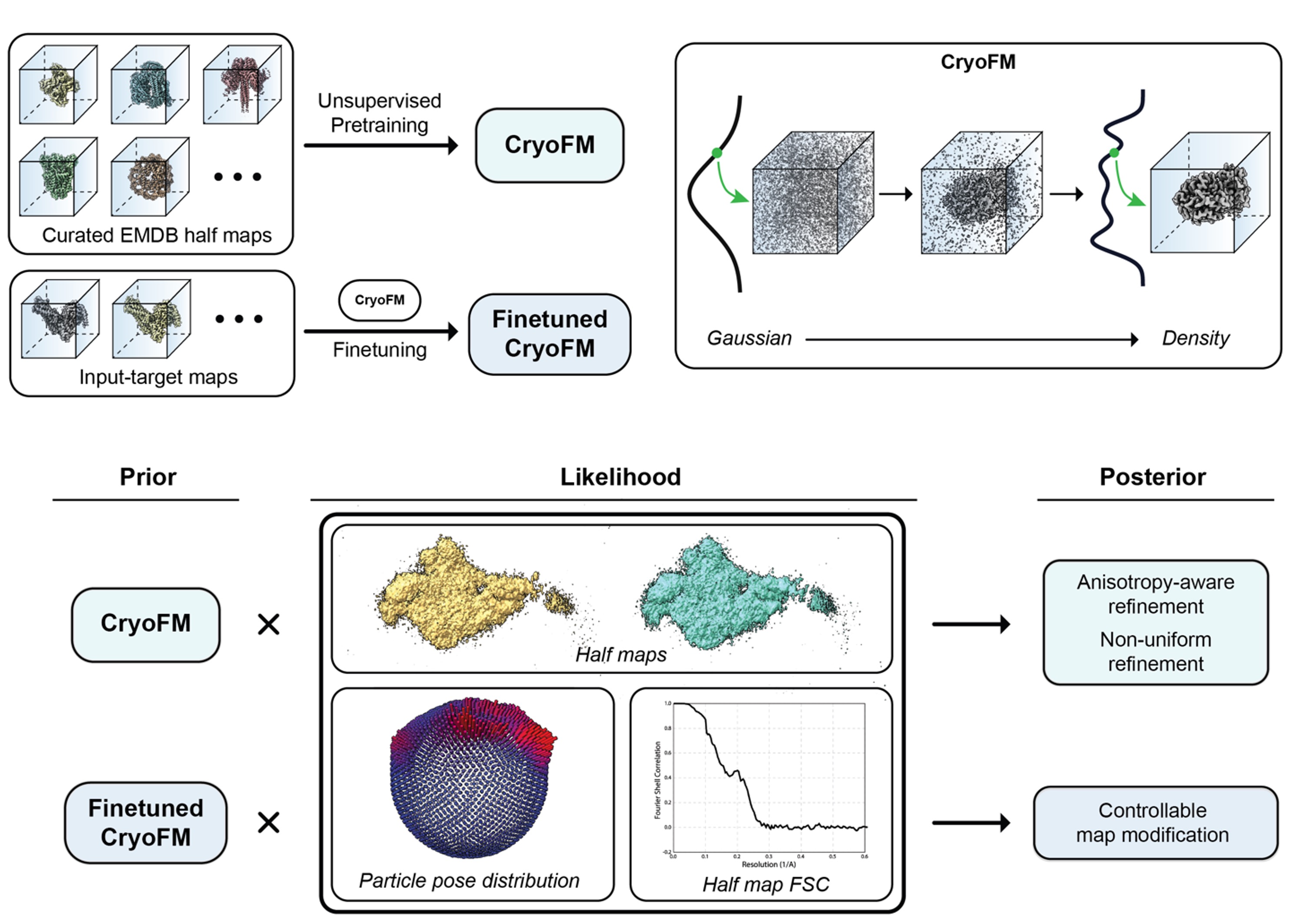
Single-particle cryo-EM density reconstruction is a severely ill-posed inverse problem, commonly degraded by noise, preferred orientations, and reconstruction artifacts. Existing approaches either rely on strong hand-crafted assumptions or supervised post-processing models.
CryoFM2 addresses this challenge by introducing a generative foundation model for cryo-EM densities. The model is trained unsupervised on thousands of high-quality cryo-EM maps using flow matching, learning a reusable prior over macromolecular density distributions that generalizes across molecular systems. The model employs a UNet architecture and is pretrained on curated EMDB half maps.
At inference time, this learned prior is combined with explicit likelihood models that describe experimental degradations, forming a principled Bayesian inference framework. Within this framework, CryoFM2 performs flow-based posterior sampling (FPS), an inference-only procedure that simultaneously enables denoising, restoration, and refinement while remaining explicitly constrained by dataset-derived statistics. This approach supports applications such as:
- Anisotropy-aware refinement
- Non-uniform reconstruction
- Controlled density modification
- Density map enhancement
Model Variants
CryoFM2 is available in three variants:
- cryofm2-pretrain: Unconditional pretrained model for general density map generation and modification tasks.
- cryofm2-emhancer: Fine-tuned model for density map enhancement (EMhancer style).
- cryofm2-emready: Fine-tuned model for density map enhancement (EMReady style).
Get Started
For End Users
- Quick Start: Learn how to use CryoFM2 for common tasks like denoising and style enhancement using simple command-line tools.
Understanding Operators
- Operators: Explore the different forward operators available in CryoFM2, including denoising, inpainting, and non-uniform refinement.
For Developers
- Unconditional Sampling: Learn how to use the Python API to generate unconditional samples from CryoFM2 models.
- Likelihood Control: Understand how to use scripts for likelihood control (FPS) and fine-tune parameters for different tasks.
- GUI Demo: Try CryoFM2 through an interactive web-based graphical interface.
Resources
- Model Weights: Available on Hugging Face.
- Source Code: Available on GitHub.
- Dataset (EMDB ID Lists): Available on Zenodo.
Citation
If you use CryoFM2 in your research, please cite:
@article{
Li2025.12.29.696802,
author={Li, Yilai and Yuan, Jing and Zhou, Yi and Wang, Zhenghua and Chen, Suyi and Yang, Fengyu and Ling, Haibin and Kovalsky, Shahar Z and Zheng, Xiaoqing and Gu, Quanquan},
title={A Generative Foundation Model for Cryo-EM Densities},
elocation-id={2025.12.29.696802},
year={2025},
doi={10.64898/2025.12.29.696802},
publisher={Cold Spring Harbor Laboratory},
URL={https://www.biorxiv.org/content/early/2025/12/29/2025.12.29.696802},
eprint={https://www.biorxiv.org/content/early/2025/12/29/2025.12.29.696802.full.pdf},
journal={bioRxiv}
}